Bioinformatics
We provide complete “END to END” solutions for next-generation sequencing and microarray on various platforms.
Selected publications using microarray technologies.
Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Cassalloa G, Scherer SW. Gene and miRNA Expression Profiles in Autism Spectrum Disorders. Brain Res. 2011 Mar 22;1380:85-97. PMID: 20868653
Bussey K, Chin K, Lababidi S, Reimers M, Reinhold WC, Kuo W, Gwadry FG, Ajay, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 Cell Line Panel. Mol Cancer Ther, 2006 Apr; 5(4):853-67. PMID: 16648555
Lee JK, Bussey KJ, Gwadry FG, Reinhold W, Riddick G, Pelletier SL, Nishizuka S, Szakacs G, Annereau JP, Shankavaram U, Lababidi S, Smith LH, Gottesman MM, Weinstein JN. Comparing cDNA and oligonucleotide array data: concordance of gene expression across platforms for the NCI-60 cancer cells. Genome Biol. 2003;4(12):R82. PMID: 14659019
Nishizuka S, Chen ST, Gwadry FG, Alexander J, Major SM, Scherf U, Reinhold WC, Waltham M, Charboneau L, Young L, Bussey KJ, Kim S, Lababidi S, Lee JK, Pittaluga S, Scudiero DA, Sausville EA, Munson PJ, Petricoin EF 3rd, Liotta LA, Hewitt SM, Raffeld M, Weinstein JN. Diagnostic markers that distinguish colon and ovarian adenocarcinomas: identification by genomic, proteomic, and tissue array profiling. Cancer Res. 2003 Sep 1;63(17):5243-50. PMID: 14500354
Upcoming publication on next-generation sequencing technologies.
Renaud Y, Gwadry FG, Daghrach A, Lemieux Perreault LP, Langlois M, Mailloux L, Mongrain I, Blazejczyk M, Dubé MP. Contribution of alignment, post-alignment and variant-calling procedures on the accuracy of next-generation sequencing pipelines (submitted).
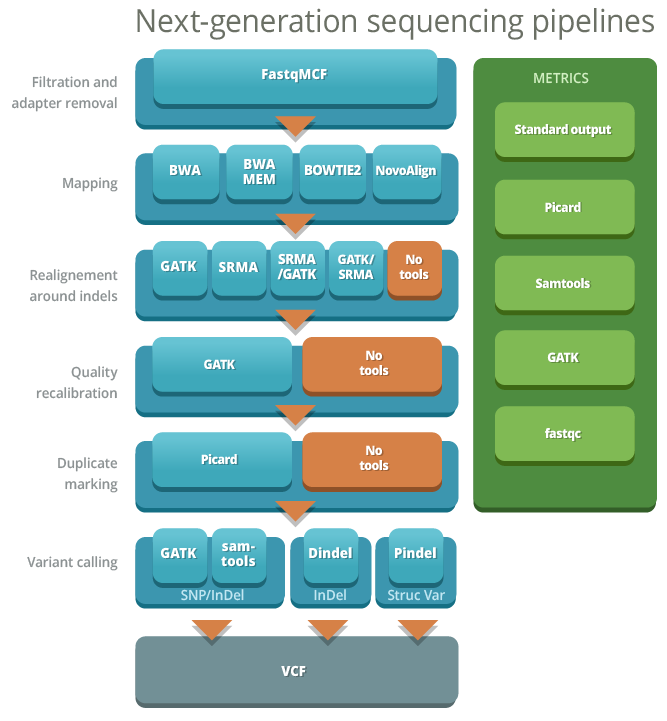
Next-generation sequencing (NGS) is a most effective way to screen for single nucleotide variants (SNVs) and small insertions and deletions (INDELs). The accurate detection of these variants is essential for clinical and research applications of NGS. One of the major challenges of NGS is the bioinformatics bottleneck for read alignment to the reference genome, postalignment processing and variant-calling. There are multiple tools for each of the steps and each have their strengths and weaknesses which can affect the quality of SNV calling. Depending on the sequencing technology the choice of tools and steps will impact the accuracy of variant calls.
Chemoinformatics
Netex Life Sciences assisted in the development of Cyclica’s (http://cyclicarx.com/) novel in-silico drug testing platform. Their platform accesses biological and chemical databases that help researchers anticipate and predict a drug candidate’s side effects in the early stages of drug development.
In addition to software development, Netex Life Sciences built a High Performance Computer cluster that allowed Cyclica to greatly improve and control their processes.